|
rbmatlab
1.16.09
|
Reduced data implementation for linear evolution problems with finite volume discretizations.
See [HO08a] Reduced Basis Method for Finite Volume Approximations of Parametrized Linear Evolution Equations for details on the implementation of the reduced matrices and vectors.
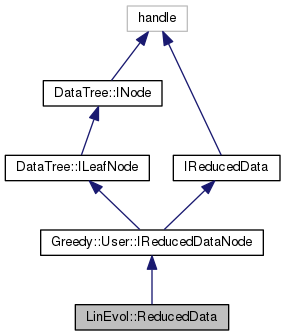
Definition at line 18 of file ReducedData.m.
Public Member Functions | |
| ReducedData (LinEvol.ReducedModel rmodel,IDetailedData detailed_data) | |
| Constructor for the generation of the reduced matrices and vectors. More... | |
| function yesno = | needs_subset_copy (LinEvol.ReducedModel rmodel) |
| function conds = | get_conds () |
 Public Member Functions inherited from Greedy.User.IReducedDataNode Public Member Functions inherited from Greedy.User.IReducedDataNode | |
| function
subset_reduced_data = | extract_reduced_data_subset (Greedy.User.IReducedModel rmodel) |
| Changes the size of the offline data matrices. More... | |
 Public Member Functions inherited from DataTree.ILeafNode Public Member Functions inherited from DataTree.ILeafNode | |
| function
children = | length () |
| Returns the number of children of the node. More... | |
| function index = | get_index (dummy2, dummy3, dummy) |
| Obtains the leaf index vector that best fits the child description given by the three arguments. More... | |
| function this = | set (dummy2,DataTree.INode value) |
| Sets a child at the given path in the tree hierarchy. More... | |
| function data = | get (index) |
| Access to a child of the current node. More... | |
| function DataTree.ILeafNode node = | get_active_leaf (DataTree.IModel model, id) |
| retuns the leaf element for the current IDetailedModel configuration. More... | |
| function tree = | create_tree (creator, ids, mu_cube, tslice, basepath) |
| Creates a new tree from a subtree specified by ids, parameter and time index regions. More... | |
 Public Member Functions inherited from DataTree.INode Public Member Functions inherited from DataTree.INode | |
| virtual function DataTree.INode tree = | create_tree (DataTree.ICreator creator, ids, mu_cube, tslice, basepath) |
| Creates a new tree from a subtree specified by ids, parameter and time index regions. More... | |
| virtual function this = | set (index, value) |
| Sets a child at the given path in the tree hierarchy. More... | |
| function | leaf_func (funcptr, ids, mu_cube, tslice) |
| applies a function to all leafs of a DataTree More... | |
| function tree = | create_scalar_tree (funcptr, ids, mu_cube, tslice) |
| copies the current trees with different leafs. These leafs are computes by a function returning scalar values. More... | |
| function
start_index = | traverse_start () |
| Start iterator for a full traverse of the DataTree. More... | |
| function
next_index = | traverse_next (this_index) |
| iterator for a full traverse of the DataTree. More... | |
| function
description = | get_active_leaf_description (IModel model, ids) |
| returns an enumeration of all leaves' basepath index vectors with a description of their parents. More... | |
| function
description = | get_leaf_description (ids, mu_cube, tslice) |
| returns an enumeration of all leaves' basepath index vectors with a description of their parents. More... | |
| function tstop = | index_valid_till (index) |
| Returns the last valid time step index of a time slice. More... | |
| function INode data = | get_by_description (id, mu, nt) |
| A combination of get_index() and get() More... | |
| function
active_leaf_index = | get_active_leaf_index (IModel model, id) |
| retuns the leaf element index for the current IDetailedModel configuration. More... | |
| function
DataTree.LeafNode active_leaf = | get_active_leaf (IModel model, id) |
| retuns the leaf element for the current IDetailedModel configuration. More... | |
| function | display (fn, basepath, name) |
| overwrites the standard display method for DataTree objects More... | |
| function lines = | disp_node (basepath, fn) |
| returns a cell array of strings with information on the node. More... | |
 Public Member Functions inherited from IReducedData Public Member Functions inherited from IReducedData | |
| virtual function
IReducedData subset_reduced_data = | extract_reduced_data_subset (rmodel) |
| Changes the size of the offline data matrices. More... | |
Static Public Member Functions | |
| static function [ LL_I , LL_E , bb , K_II , K_IE , K_EE , m_I , m_E , m ] = | rb_operators (LinEvol.ReducedModel rmodel,Greedy.DataTree.Detailed.RBLeafNode detailed_data, decomp_mode) |
| function computing the time-dependent reduced basis operators and vectors. More... | |
Public Attributes | |
| a0 | |
| Dof vectors of projections \(\left\{{ \cal P }_{\text{red}}[u_0^q] \right\}_{q=1}^{Q_{u_0}}\). More... | |
| LL_I | |
| \(Q_I\) reduced matrix for implicit operator \({\cal L}_{h,I}\) More... | |
| LL_E | |
| \(Q_E\)-sequence of reduced matrices for explicit operator \({\cal L}_{h,E}\) More... | |
| bb | |
| \(Q_b\)-sequence of reduced vectors for constant contributions \(b_h\) More... | |
| K_II | |
| \(Q_I \cdot Q_I\)-sequence of gram matrices More... | |
| K_IE | |
| \(Q_I \cdot Q_E\)-sequence of gram matrices More... | |
| K_EE | |
| \(Q_E \cdot Q_E\)-sequence of gram matrices More... | |
| m_I | |
| \(Q_I \cdot Q_b\)-sequence of vectors More... | |
| m_E | |
| \(Q_E \cdot Q_b\)-sequence of vectors More... | |
| m | |
| \(Q_b \cdot Q_b\)-sequence of scalars More... | |
| s_RB | |
| in case of output functional this is a \(Q_s\)-sequence of vectors. More... | |
| s_l2norm | |
| in case of output functional this is a \(Q_s\)-sequence of scalars. More... | |
| N | |
| number of reduced basis vectors stored in this data node. More... | |
| M = 0 | |
| number of collateral reduced basis vectors stored in this data node. More... | |
| Mstrich = 0 | |
| number of collateral reduced basis vectors used for error estimation More... | |
 Public Attributes inherited from Greedy.User.IReducedDataNode Public Attributes inherited from Greedy.User.IReducedDataNode | |
| methods *function | rdn |
| LinEvol.ReducedData.ReducedData | ( | LinEvol.ReducedModel | rmodel, |
| IDetailedData | detailed_data | ||
| ) |
Constructor for the generation of the reduced matrices and vectors.
| rmodel | object specifying how the reduced simulations can be computed. |
| detailed_data | object defining the basis generation algorithm and storage for storing high dimensional data, i.e. dependent on dimension \(H\). This data is necessary for detailed simulations, construction of online matrices, reduced_data and reconstruction of reduced simulations. |
Definition at line 219 of file ReducedData.m.
| function yesno = LinEvol.ReducedData.needs_subset_copy | ( | LinEvol.ReducedModel | rmodel | ) |
Definition at line 261 of file ReducedData.m.
|
static |
function computing the time-dependent reduced basis operators and vectors.
In coefficients mode the detailed data is empty.
| rmodel | object specifying how the reduced simulations can be computed. |
| detailed_data | object defining the basis generation algorithm and storage for storing high dimensional data, i.e. dependent on dimension \(H\). This data is necessary for detailed simulations, construction of online matrices, reduced_data and reconstruction of reduced simulations. |
| decomp_mode | flag indicating the operation mode of the function:
|
| LL_I | \(Q_I\) reduced matrix for implicit operator \({\cal L}_{h,I}\) |
| LL_E | \(Q_E\)-sequence of reduced matrices for explicit operator \({\cal L}_{h,E}\) |
| bb | \(Q_b\)-sequence of reduced vectors for constant contributions \(b_h\) |
| K_II | \(Q_I \cdot Q_I\)-sequence of gram matrices |
| K_IE | \(Q_I \cdot Q_E\)-sequence of gram matrices |
| K_EE | \(Q_E \cdot Q_E\)-sequence of gram matrices |
| m_I | \(Q_I \cdot Q_b\)-sequence of vectors |
| m_E | \(Q_E \cdot Q_b\)-sequence of vectors |
| m | \(Q_b \cdot Q_b\)-sequence of scalars |
descr — descrmodel_data — model data RB — RB Definition at line 19 of file rb_operators.m.
| LinEvol.ReducedData.a0 |
Dof vectors of projections \(\left\{{ \cal P }_{\text{red}}[u_0^q] \right\}_{q=1}^{Q_{u_0}}\).
Definition at line 34 of file ReducedData.m.
| LinEvol.ReducedData.bb |
\(Q_b\)-sequence of reduced vectors for constant contributions \(b_h\)
\[({ \bf b})_{i} = \int \varphi_i b^q_h\]
for \(i=1,\ldots,N\) and \(q=1,\ldots,Q_b\).Definition at line 69 of file ReducedData.m.
| LinEvol.ReducedData.K_EE |
\(Q_E \cdot Q_E\)-sequence of gram matrices
\[({ \bf K}_{EE}^{q_1,q_2})_{ij} = \int {\cal L}^{q_1}_{h,E}[\varphi_i] {\cal L}^{q_2}_{h,E}[\varphi_j]\]
for \(i,j=1,\ldots,N\) and \(q_1,q_2=1,\ldots,Q_E\).Definition at line 105 of file ReducedData.m.
| LinEvol.ReducedData.K_IE |
\(Q_I \cdot Q_E\)-sequence of gram matrices
\[({ \bf K}_{IE}^{q_1,q_2})_{ij} = \int {\cal L}^{q_1}_{h,I}[\varphi_i] {\cal L}^{q_2}_{h,E}[\varphi_j]\]
for \(i,j=1,\ldots,N\) and \(q_1=1,\ldots,Q_I, q_2=1,\ldots,Q_E\).Definition at line 93 of file ReducedData.m.
| LinEvol.ReducedData.K_II |
\(Q_I \cdot Q_I\)-sequence of gram matrices
\[({ \bf K}_{II}^{q_1,q_2})_{ij} = \int {\cal L}^{q_1}_{h,I}[\varphi_i] {\cal L}^{q_2}_{h,I}[\varphi_j]\]
for \(i,j=1,\ldots,N\) and \(q_1,q_2=1,\ldots,Q_I\).Definition at line 81 of file ReducedData.m.
| LinEvol.ReducedData.LL_E |
\(Q_E\)-sequence of reduced matrices for explicit operator \({\cal L}_{h,E}\)
\[({ \bf L}_E^q)_{ij} = \int \varphi_i {\cal L}^q_{h,E}[\varphi_j]\]
for \(i,j=1,\ldots,N\) and \(q=1,\ldots,Q_E\).Definition at line 57 of file ReducedData.m.
| LinEvol.ReducedData.LL_I |
\(Q_I\) reduced matrix for implicit operator \({\cal L}_{h,I}\)
\[({ \bf L}_I^q)_{ij} = \int \varphi_i {\cal L}^q_{h,I}[\varphi_j]\]
for \(i,j=1,\ldots,N\) and \(q=1,\ldots,Q_I\).Definition at line 45 of file ReducedData.m.
| LinEvol.ReducedData.m |
\(Q_b \cdot Q_b\)-sequence of scalars
\[{ \bf m}^{q_1,q_2} = \int b_h^{q_1} b_h^{q_2}\]
and \(q_1,q_2=1,\ldots,Q_b\).Definition at line 141 of file ReducedData.m.
| LinEvol.ReducedData.M = 0 |
number of collateral reduced basis vectors stored in this data node.
SetAccess = Private, GetAccess = Public Definition at line 193 of file ReducedData.m.
| LinEvol.ReducedData.m_E |
\(Q_E \cdot Q_b\)-sequence of vectors
\[({ \bf m}_E^{q_1,q_2})_i = \int {\cal L}^{q_1}_{h,E}[\varphi_i] b_h^{q_2}\]
for \(i=1,\ldots,N\) and \(q_1=1,\ldots,Q_E, q_2=1,\ldots,Q_b\).Definition at line 129 of file ReducedData.m.
| LinEvol.ReducedData.m_I |
\(Q_I \cdot Q_b\)-sequence of vectors
\[({ \bf m}_I^{q_1,q_2})_i = \int {\cal L}^{q_1}_{h,I}[\varphi_i] b_h^{q_2}\]
for \(i=1,\ldots,N\) and \(q_1=1,\ldots,Q_I, q_2=1,\ldots,Q_b\).Definition at line 117 of file ReducedData.m.
| LinEvol.ReducedData.Mstrich = 0 |
number of collateral reduced basis vectors used for error estimation
SetAccess = Private, GetAccess = Public Definition at line 204 of file ReducedData.m.
| LinEvol.ReducedData.N |
number of reduced basis vectors stored in this data node.
Dependent set to true. SetAccess = Private, GetAccess = Public Definition at line 179 of file ReducedData.m.
| LinEvol.ReducedData.s_l2norm |
in case of output functional this is a \(Q_s\)-sequence of scalars.
\[{ \bf s}_{l^2}^q = \|s(\varphi_1), \ldots, s(\varphi_N)\|_{l^2}\]
for \(q_1=1,\ldots,Q_s\)Definition at line 165 of file ReducedData.m.
| LinEvol.ReducedData.s_RB |
in case of output functional this is a \(Q_s\)-sequence of vectors.
\[({ \bf s}^q)_i = s(\varphi_i)\]
for \(i=1,\ldots,N\) and \(q_1=1,\ldots,Q_s\)Definition at line 153 of file ReducedData.m.
 1.8.8
1.8.8