|
rbmatlab
1.16.09
|
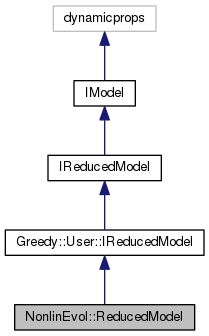
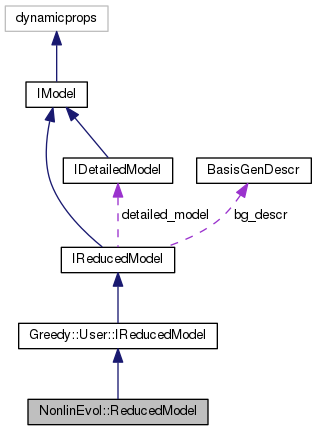
reduced model for non-linear evolution problems as given by a NonlinEvol.DetailedModel.
This is compatible with Greedy.User.IReducedModel and can therefore make use of detailed data objects created by Greedy algorithms.
Definition at line 18 of file ReducedModel.m.
Public Member Functions | |
| ReducedModel (NonlinEvol.DetailedModel dmodel,BasisGenDescr bg_descr) | |
| Constructor for the reduced model. More... | |
| function M = | get_current_M (id) |
| returns the collateral reduced basis size for the current parameter and time instant More... | |
| function NonlinEvol.ReducedModel c = | copy () |
| function that deep copies this handle class More... | |
| function
rb_sim_data = | rb_simulation_impl (NonlinEvol.EiRbReducedDataNode reduced_data) |
| function, which performs a reduced basis online simulation for the parameter vector \(\mu \in {\cal P} \subset \mathbb{R}^p\), which is assumed to be set by IDetailedModel.set_mu() More... | |
| function a0 = | rb_init_values (Greedy.DataTree.Detailed.PODEILeafNode detailed_data, decomp_mode) |
| function computing initial values for a reduced simulation. More... | |
| function
rb_sim_data = | rb_reconstruction (NonlinEvol.DetailedData detailed_data, rb_sim_data) |
| (trivial) function computing a detailed reconstruction by linear combination of the coefficients in the simulation data with the orthonormal reduced basis RB More... | |
 Public Member Functions inherited from Greedy.User.IReducedModel Public Member Functions inherited from Greedy.User.IReducedModel | |
| IReducedModel (IDetailedModel dmodel,BasisGenDescr bg_descr) | |
| constructor for this reduced model interface More... | |
| function
Greedy.User.ReducedData reduced_data = | gen_reduced_data (Greedy.User.IDetailedData detailed_data) |
Constructs the reduced_data object holding low dimensional data needed for efficient reduced simulations with rb_simulation(). More... | |
| function
rb_sim_data = | rb_simulation (Greedy.User.ReducedData reduced_data) |
| forwards the reduced simulation to the method rb_simulation_impl() after getting a suitable reduced data leaf element. More... | |
| virtual function
rb_sim_data = | rb_simulation_impl (Greedy.User.IReducedDataNode reduced_data) |
| implementation of the reduced simulation More... | |
 Public Member Functions inherited from IReducedModel Public Member Functions inherited from IReducedModel | |
| IReducedModel (IDetailedModel dmodel, bg_descr) | |
| Constructor of a reduced model. More... | |
| function iseq = | eq (IReducedModel other) |
Comparison operator checking whether the underlying detailed_model members of this and other are equal. More... | |
| function IReducedData reduced_data = | gen_reduced_data (detailed_data) |
Constructs the reduced_data object holding low dimensional data needed for efficient reduced simulations with rb_simulation(). More... | |
| function
reduced_data_subset = | extract_reduced_data_subset (IReducedData reduced_data) |
Extracts a subset of the reduced_data generated by gen_reduced_data(). More... | |
| virtual function
rb_sim_data = | rb_simulation (reduced_data) |
| Executes a reduced simulation and optionally an error estimation. More... | |
| virtual function
rb_sim_data = | rb_reconstruction (detailed_data, rb_sim_data) |
| reconstructs the reduced simulation snapshots generated by rb_simulation() in the reduced space \({\cal W}_{\text{red}}\). More... | |
| function U = | get_dofs_from_sim_data (sim_data) |
extracts the \(H\) dimensional Dof vector from the sim_data structure More... | |
| function IDetailedData detailed_data = | gen_detailed_data (model_data) |
| initiates the reduced basis generation process More... | |
| function p = | plot_sim_data (model_data, sim_data, plot_params) |
| plots the simulation data as returned by detailed_simulation() More... | |
| function
model_data = | gen_model_data () |
| generates large model data. More... | |
| function
sim_data = | detailed_simulation (model_data) |
| executes a detailed simulation for a given parameter More... | |
| function this = | set_mu (mu) |
| Sets the active parameter vector \(\mu \in {\cal M}\) used for simulations on this model. More... | |
| function mu = | get_mu () |
| returns the active parameter vector \(\mu \in { \cal M }\) More... | |
| function
rb_size = | get_rb_size (detailed_data) |
| returns the size of the generated reduced basis by the IDetailedData class. More... | |
| function | couple_N_and_M (detailed_data, ratio, factor) |
| sets all the basis sizes for reduced simulations by a ratio with respect to the maximum possible basis size and a factor between RB and EI basis sizes. More... | |
| function this = | set_Mratio (detailed_data, ratio) |
| in case of multiple operators subject to empirical interpolation, this sets number of reduced basis functions used for reduced simulations by specifying a ratio. More... | |
| function Mratio = | get_Mratio (detailed_data) |
| in case of multiple operators subject to empirical interpolation, this gets the mean of ratio between the number of reduced basis functions used for reduced simulations and the maximum possible. More... | |
| function
varargout = | subsref (S) |
| forwarding of fieldnames access to the underlying detailed_model description More... | |
Static Public Member Functions | |
| static function Delta = | get_estimators_from_sim_data (rb_sim_data) |
| static function Delta = | get_estimator_from_sim_data (rb_sim_data) |
| Static helper method returning an error estimator for the whole reduced trajectory \(\{u_{\text{red}}(\cdot, t^k)\}_{k=0}^{K}\) generated by rb_simulation(). More... | |
 Static Public Member Functions inherited from IModel Static Public Member Functions inherited from IModel | |
| static function ok = | struct_check (descr, checks) |
| executes checks on the fields of a structure object More... | |
Public Attributes | |
| enable_error_estimator = true | |
| compute_conditions = false | |
| properties (Constant) More... | |
| stencil_mode = "edge" | |
| defines how the stencil of the local grid is computed More... | |
| local_stencil_size = 1 | |
| defines the number of neighbors that shall be added to the stencil of the local grid created for efficient empirical interpolation evalutations. More... | |
| t | |
| current time instance More... | |
| tstep | |
| current time step More... | |
 Public Attributes inherited from IReducedModel Public Attributes inherited from IReducedModel | |
| descr | |
| The description structure holding information about the analytical parametrized problem and its discretization. More... | |
| decomp_mode | |
| mu_names | |
| cell array of strings describing the parameters of the model More... | |
| mu_ranges | |
| cell array of vectors of size two defining the allowed interval range for the parameter components More... | |
| verbose | |
| an integer defining the verbosity level of information output during basis generation More... | |
| debug | |
| an integer defining the debugging level controlling error output and extra tests during basis generation More... | |
| crb_enabled = false | |
| flag indicating whether this model depends on collateral reduced basis spaces. More... | |
| ::IDetailedModel | detailed_model |
| an object which shall be reduced | |
| ::BasisGenDescr | bg_descr |
| a structure defining the basis generation routines and data structures. | |
| enable_error_estimator | |
| boolean flag indicating whether during an rb_simulation() an a posteriori error estimator shall be computed. More... | |
| N = 0 | |
| control variable for the size of the reduced basis used for reduced simulations. By default this is equal to the size of the generated reduced basis. More... | |
| M = 0 | |
| control variable for the size of the (collateral) reduced basis used for empirical interpolations. By default this is equal to the size of the generated reduced basis. More... | |
| Mstrich = 0 | |
| control variable for the number of (collateral) reduced basis vectors used for error estimation. By default this is equal to zero. More... | |
 Public Attributes inherited from IModel Public Attributes inherited from IModel | |
| num_cpus = 4 | |
| The number of CPUs used for parallel sessions. More... | |
| decomp_mode | |
| Decomposition operation mode. More... | |
| mu_names | |
| cell array of strings describing the parameters of the model More... | |
| mu_ranges | |
| cell array of vectors of size two defining the allowed interval range for the parameter components More... | |
| verbose | |
| an integer defining the verbosity level of information output during basis generation More... | |
| debug | |
| an integer defining the debugging level controlling error output and extra tests during basis generation More... | |
Static Public Attributes | |
| static const | ddescr_checks |
| This constant is used for a consistency check of the model descr with help of IModel.struct_check() More... | |
 Static Public Attributes inherited from IModel Static Public Attributes inherited from IModel | |
| static const | time_checks |
| This constant can be used for a consistency check of time evolution members in the ModelDescr with help of IModel.struct_check() More... | |
| NonlinEvol.ReducedModel.ReducedModel | ( | NonlinEvol.DetailedModel | dmodel, |
| BasisGenDescr | bg_descr | ||
| ) |
Constructor for the reduced model.
| dmodel | object specifying how the high dimensional data can be computed. |
| bg_descr | structure specifying how the reduced basis shall be generated. |
descr — descr Definition at line 130 of file ReducedModel.m.
|
virtual |
function that deep copies this handle class
| c | an object which is a deep copy of this object. |
Implements IReducedModel.
Definition at line 194 of file ReducedModel.m.
| function M = NonlinEvol.ReducedModel.get_current_M | ( | id | ) |
returns the collateral reduced basis size for the current parameter and time instant
| id | an optional id in case there are many collateral reduced basis spaces for one parameter and time instant combination. |
| M | the size of the collateral reduced basis space |
Definition at line 168 of file ReducedModel.m.
|
staticvirtual |
Static helper method returning an error estimator for the whole reduced trajectory \(\{u_{\text{red}}(\cdot, t^k)\}_{k=0}^{K}\) generated by rb_simulation().
| rb_sim_data | struct holding reduced simulation data returned by IReducedModel.rb_simulation() . struct holding reduced simulation data returned by IReducedModel.rb_simulation() . |
| Delta | This is a (K+1) x 1 vector of estimates \(\eta^k(\mu)\) Delta = get_estimators_from_sim_data(rb_sim_data); This is a scalar computed from the estimates \(\eta^k(\mu)\). Usually the maximum over \(k=0,\ldots,K\) is returned. |
Implements IReducedModel.
Definition at line 290 of file ReducedModel.m.
|
static |
Definition at line 279 of file ReducedModel.m.
| function a0 = NonlinEvol.ReducedModel.rb_init_values | ( | Greedy.DataTree.Detailed.PODEILeafNode | detailed_data, |
| decomp_mode | |||
| ) |
function computing initial values for a reduced simulation.
This calls rb_init_values_separable().
| detailed_data | detailed data tree leaf object |
| decomp_mode | flag indicating the operation mode of the function:
|
| a0 | coefficient vector of size N x 1 for the initial values. |
Definition at line 212 of file ReducedModel.m.
| function rb_sim_data = NonlinEvol.ReducedModel.rb_reconstruction | ( | NonlinEvol.DetailedData | detailed_data, |
| rb_sim_data | |||
| ) |
(trivial) function computing a detailed reconstruction by linear combination of the coefficients in the simulation data with the orthonormal reduced basis RB
This calls rb_reconstruction_default().
| detailed_data | object defining the basis generation algorithm and storage for storing high dimensional data, i.e. dependent on dimension \(H\). This data is necessary for detailed simulations, construction of online matrices, reduced_data and reconstruction of reduced simulations. |
| rb_sim_data | struct holding reduced simulation data returned by ReducedModel.rb_simulation(). |
| rb_sim_data | struct holding the reduced simulation results and their reconstructions. |
Definition at line 237 of file ReducedModel.m.
| function rb_sim_data = NonlinEvol.ReducedModel.rb_simulation_impl | ( | NonlinEvol.EiRbReducedDataNode | reduced_data | ) |
function, which performs a reduced basis online simulation for the parameter vector \(\mu \in {\cal P} \subset \mathbb{R}^p\), which is assumed to be set by IDetailedModel.set_mu()
The behaviour of this simulation is controlled by the ModelDescr structure rmodel.descr.
| reduced_data | an object constructing and storing all (low-dimensional) reduced matrices and vectors needed for reduced simulations. |
| rb_sim_data | rb sim data |
mu_names — the cell array of names of mu-components and for each of these stringt, a corresponding field in model is expected. T — end-time of simulation nt — number of timesteps to compute L_E_local_name — name of the local space-discretization operator evaluation with syntax *_local indicates results on a set of elements, *_local_ext includes information including their neighbours, i.e. the grid must be known also for the neighbours, the values of the previous timstep must be known on the neighbours. The subset of elements, which are to be computed are given in ind_local and the values produced are INC_local. Subsequent time-step operator would be defined as NU_local = U(ind_local) - dt * INC_local rb_operators — function pointer to a method assembling the decomposed discretization operatorsa0 — a0 LL_I — LL I bb_I — bb Idata_const_in_time — if this flag is set, then only operators for first time instance are computeda — a(:,k) is the coefficient vector of the reduced simulation at time \(t^{k-1}\) Delta — Delta reslog — reslog eilog — eilog conds.LI — conds.LI conds.gradLI — conds.gradLI Definition at line 19 of file rb_simulation_impl.m.
| NonlinEvol.ReducedModel.compute_conditions = false |
properties (Constant)
copybrief NonlinEvol.DetailedModel.compute_conditions
Default: false
Definition at line 38 of file ReducedModel.m.
|
static |
This constant is used for a consistency check of the model descr with help of IModel.struct_check()
Default: struct(" \ 'init_values_algorithm', {{@(x) isequal(class(x), 'function_handle')}}, \ 'error_norm', {{@ischar, 'values', {'l2', 'energy'} }}, \ 'stencil_mode', {{@ischar, 'values', {'edge', 'vertex'} }}, \ 'local_stencil_size', {{@isnumeric}} ")
Definition at line 107 of file ReducedModel.m.
| NonlinEvol.ReducedModel.enable_error_estimator = true |
Definition at line 34 of file ReducedModel.m.
| NonlinEvol.ReducedModel.local_stencil_size = 1 |
defines the number of neighbors that shall be added to the stencil of the local grid created for efficient empirical interpolation evalutations.
Default: 1
Definition at line 69 of file ReducedModel.m.
| NonlinEvol.ReducedModel.stencil_mode = "edge" |
defines how the stencil of the local grid is computed
edge for all neighbours over edgesvertex for all neighbours of a grid cell's vertices
Default: "edge"
Definition at line 54 of file ReducedModel.m.
| NonlinEvol.ReducedModel.t |
current time instance
Dependent set to true. Definition at line 83 of file ReducedModel.m.
| NonlinEvol.ReducedModel.tstep |
current time step
Dependent set to true. Definition at line 94 of file ReducedModel.m.
 1.8.8
1.8.8