 |
KerMor
0.9
Model order reduction for nonlinear dynamical systems and nonlinear approximation
|
 |
KerMor
0.9
Model order reduction for nonlinear dynamical systems and nonlinear approximation
|
This class implements the nonlinear continuum mechanics as described in [5] . More...
This class implements the nonlinear continuum mechanics as described in [5] .
Definition at line 19 of file Dynamics.m.
Public Member Functions | |
| Dynamics (sys) | |
| function | configUpdated () |
| function | prepareSimulation (colvec< double > mu) |
| function fx = | evaluateCoreFun (colvec< double > x,double t) |
| Actual method used to evaluate the dynamical sytems' core function. More... | |
| function fx = | evaluateComponentSet (integer nr,colvec< double > x,double t) |
| Computes the full or reduced component functions of the given point set. More... | |
| function fx = | evaluateComponentSetMulti (integer nr,matrix< double > x,rowvec< double > t,matrix< double > mu) |
| Computes the full component functions of the given point set. More... | |
| function res = | test_Jacobian (matrix< double > y,double t,colvec< double > mu) |
| Overrides the random argument jacobian test as restrictions on the possible x values (detF = 1) hold. More... | |
| function copy = | clone () |
| Create new instance. More... | |
| function | setSystem (sys) |
| function copy = | project (V, W) |
| function [
SPK , SPg , SPalpha , SPLamDot ] = | computeSparsityPattern () |
| Computes all sorts of patterns simultaneously. More... | |
 Public Member Functions inherited from dscomponents.ACompEvalCoreFun Public Member Functions inherited from dscomponents.ACompEvalCoreFun | |
| ACompEvalCoreFun (sys) | |
| function fx = | evaluateComponentSet (integer nr,colvec< double > x,double t) |
| Computes the full or reduced component functions of the given point set. More... | |
| function fx = | evaluateComponentSetMulti (integer nr,matrix< double > x,rowvec< double > t,matrix< double > mu) |
| Computes the full component functions of the given point set. More... | |
| function dfx = | evaluateComponentSetGradientsAt (integer nr,colvec< double > x,double t) |
| Computes the full/reduced gradients of all component functions of the given point set. More... | |
| function J = | evaluateJacobianSet (integer nr,colvec< double > x,double t) |
| Returns the jacobian entries of the point set that have been specified using setPointSet's argument jpd. More... | |
| function J = | evaluateJacobianSetMulti (integer nr,matrix< double > x,rowvec< double > t,colvec< double > mu) |
| Returns the jacobian entries at multiple locations/times/parameters of the point set that have been specified using setPointSet's argument jpd. More... | |
| function | setPointSet (nr, pts, jpd) |
| Parameters: pts: A row vector with the desired points jpd: ("Jacobian Partial Derivatives") A cell array of size equal to the number of points. Each cell contains the indices for which the partial derivatives of the corresponding component function will be computed when calling evaluateJacobianSet. More... | |
| function target = | project (V, W, target) |
| function copy = | clone (copy) |
| The interface method with returns a copy of the current class instance. More... | |
| function res = | test_ComponentEvalMatch (xsize) |
| Tests if the local implementation of evaluateComponents matches the full evaluation. More... | |
 Public Member Functions inherited from dscomponents.ACoreFun Public Member Functions inherited from dscomponents.ACoreFun | |
| ACoreFun (sys) | |
| function | setSystem (sys) |
| function target = | project (V, W, target) |
| Sets the protected \(\vV,\vW\) matrices that can be utilized on core function evaluations after projection. More... | |
| function fx = | evaluate (x, t) |
| Evaluates the f-approximation. Depending on a possible projection and the CustomProjection-property the function either calls the inner evaluation directly which assumes \(f = f^r(z)\) or projects the reduced state variable z into the original space and evaluates the function there, so via \(f = V'f(Vz)\). More... | |
| function fx = | evaluateMulti (colvec< double > x,double t,colvec< double > mu) |
| Evaluates this function on multiple locations and maybe multiple times and parameters. More... | |
| function | prepareSimulation (colvec< double > mu) |
| A method that allows parameter-dependent computations to be performed before a simulation using this parameter starts. More... | |
| function J = | getStateJacobian (x, t) |
| Default implementation of jacobian matrix evaluation via finite differences. More... | |
| function J = | getStateJacobianImpl (colvec< double > x,double t) |
| Default implementation of state jacobians. uses finite differences. More... | |
| function copy = | clone (copy) |
| The interface method with returns a copy of the current class instance. More... | |
| function [
matrix< double > J , dx ] = | getStateJacobianFD (x, t,rowvec< integer > partidx) |
| Implementation of jacobian matrix evaluation via finite differences. More... | |
| function res = | test_MultiArgEval (mudim) |
| Convenience function that tests if a custom MultiArgumentEvaluation works as if called with single arguments. More... | |
| function logical res = | test_Jacobian (matrix< double > xa,rowvec< double > ta,matrix< double > mua) |
| Tests the custom provided jacobian matrix against the default finite difference computed one. More... | |
 Public Member Functions inherited from KerMorObject Public Member Functions inherited from KerMorObject | |
| KerMorObject () | |
| Constructs a new KerMor object. More... | |
| function | display () |
| disp(object2str(this)); More... | |
| function bool = | eq (B) |
| Checks equality of two KerMor objects. More... | |
| function bool = | ne (B) |
| Checks if two KerMorObjects are different. More... | |
| function cn = | getClassName () |
| Returns the simple class name of this object without packages. More... | |
 Public Member Functions inherited from DPCMObject Public Member Functions inherited from DPCMObject | |
| DPCMObject () | |
| Creates a new DPCM object. More... | |
| DPCMObject () | |
 Public Member Functions inherited from general.AProjectable Public Member Functions inherited from general.AProjectable | |
| function handle target = | project (matrix< double > V,matrix< double > W,handle target) |
| Returns a NEW INSTANCE of the projected object that does not rely on data of the old one via references (everything must be copied to ensure separability of reduced(=projected) versions and full versions, unless. More... | |
| function copy = | clone (copy) |
| The interface method with returns a copy of the current class instance. More... | |
Public Attributes | |
| b1cf = 5.316372204148964 | |
| Cross-fibre markert part [MPa] = [N/mm²]. More... | |
| d1cf = 0.014991843974911 | |
| [-] More... | |
| function_handle | alpha = @("t")0 |
| The activation of the muscle at time t [-]. More... | |
| ComputeUnassembled = false | |
| % Unassembled stuff More... | |
| Sigma | |
| Sigma assembly matrix. More... | |
| idx_uv_bc_glob_unass | |
| The indices of any dirichlet value in the unassembled vector duvw. More... | |
| fDim_unass | |
| num_dv_unass | |
| idx_v_elems_unass | |
| ForceLengthFun | |
| % Force length function fields More... | |
| ForceLengthFunDeriv | |
| APExp | |
| AnisoPassiveMuscle | |
| AnisoPassiveMuscleDeriv | |
| AnisoPassiveTendon | |
| AnisoPassiveTendonDeriv | |
| lambda_dot_pos | |
| Helper variable for fullmodels.muscle.model. More... | |
| lambda_dot | |
| nfibres | |
| LastBCResiduals | |
| Prepared arguments for APExpansion muprep;. More... | |
| nfevals | |
| nJevals | |
| RampTime | |
| Helper value for QuickReleaseTests (or others) that use a function handle with certain alpha ramp time for different simulations. Used in getSimCacheExtra to uniquely identify a simulation in the cache. More... | |
 Public Attributes inherited from dscomponents.ACompEvalCoreFun Public Attributes inherited from dscomponents.ACompEvalCoreFun | |
| PointSets | |
 Public Attributes inherited from dscomponents.ACoreFun Public Attributes inherited from dscomponents.ACoreFun | |
| logical | TimeDependent = true |
| Flag that indicates if the ACoreFun is (truly) time-dependent. More... | |
| CustomProjection = false | |
| Set this property if the projection process is customized by overriding the default project method. More... | |
| sparse< logical > | JSparsityPattern = "[]" |
| Sparsity pattern for the jacobian matrix. More... | |
| integer | xDim = "[]" |
| The current state space dimension of the function's argument \(x\). More... | |
| integer | fDim = "[]" |
| The current output dimension of the function. More... | |
| models.BaseFirstOrderSystem | System |
| The system associated with the current ACoreFun. More... | |
| colvec< double > | mu = "[]" |
| The current model parameter mu for evaluations. Will not be persisted as only valid for runtime during simulations. More... | |
| Vcache | |
| Wcache | |
 Public Attributes inherited from DPCMObject Public Attributes inherited from DPCMObject | |
| WorkspaceVariableName = "" | |
| The workspace variable name of this class. Optional. More... | |
| ID = "[]" | |
| An ID that allows to uniquely identify this DPCMObject (at least within the current MatLab session/context). More... | |
| PropertiesChanged = "[]" | |
| The Dictionary containing all the property settings as key/value pairs. More... | |
 Public Attributes inherited from handle Public Attributes inherited from handle | |
| addlistener | |
| Creates a listener for the specified event and assigns a callback function to execute when the event occurs. More... | |
| notify | |
| Broadcast a notice that a specific event is occurring on a specified handle object or array of handle objects. More... | |
| delete | |
| Handle object destructor method that is called when the object's lifecycle ends. More... | |
| disp | |
| Handle object disp method which is called by the display method. See the MATLAB disp function. More... | |
| display | |
| Handle object display method called when MATLAB software interprets an expression returning a handle object that is not terminated by a semicolon. See the MATLAB display function. More... | |
| findobj | |
| Finds objects matching the specified conditions from the input array of handle objects. More... | |
| findprop | |
| Returns a meta.property objects associated with the specified property name. More... | |
| fields | |
| Returns a cell array of string containing the names of public properties. More... | |
| fieldnames | |
| Returns a cell array of string containing the names of public properties. See the MATLAB fieldnames function. More... | |
| isvalid | |
| Returns a logical array in which elements are true if the corresponding elements in the input array are valid handles. This method is Sealed so you cannot override it in a handle subclass. More... | |
| eq | |
| Relational functions example. See details for more information. More... | |
| transpose | |
| Transposes the elements of the handle object array. More... | |
| permute | |
| Rearranges the dimensions of the handle object array. See the MATLAB permute function. More... | |
| reshape | |
| hanges the dimensions of the handle object array to the specified dimensions. See the MATLAB reshape function. More... | |
| sort | |
| ort the handle objects in any array in ascending or descending order. More... | |
 Public Attributes inherited from general.AProjectable Public Attributes inherited from general.AProjectable | |
| V | |
| The \(V\) matrix of the biorthogonal pair \(V,W\). More... | |
| W | |
| The \(W\) matrix of the biorthogonal pair \(V,W\). More... | |
Protected Member Functions | |
| function fx = | evaluateComponents (rowvec< integer > pts,rowvec< integer > ends, unused1, unused2,matrix< double > x, unused3) |
| This is the template method that actually evaluates the components at given points and values. More... | |
| function dfx = | evaluateComponentPartialDerivatives (rowvec< integer > pts,rowvec< integer > ends,rowvec< integer > idx,rowvec< integer > deriv,rowvec< integer > self,colvec< double > x,double t, dfxsel) |
| Computes specified partial derivatives of \(f\) of the components given by pts and the selected partial derivatives by dfxsel. More... | |
 Protected Member Functions inherited from dscomponents.ACompEvalCoreFun Protected Member Functions inherited from dscomponents.ACompEvalCoreFun | |
| function matrix < double > dfx = | evaluateComponentGradientsAt (rowvec< integer > pts,rowvec< integer > ends,rowvec< integer > idx,rowvec< integer > self,colvec< double > x,double t) |
| Default implementation of gradient computation via finite differences. More... | |
| function dfx = | evaluateComponentPartialDerivatives (rowvec< integer > pts,rowvec< integer > ends,rowvec< integer > idx,rowvec< integer > deriv,rowvec< integer > self,colvec< double > x,double t, dfxsel) |
| Computes specified partial derivatives of \(f\) of the components given by pts and the selected partial derivatives by dfxsel. More... | |
| function dfx = | evaluateComponentPartialDerivativesMulti (pts, ends, idx, deriv, self,colvec< double > x,double t,colvec< double > mu, dfxsel) |
| Multi-argument evaluation method for partial derivatives. Not used so far in KerMor, this is "legacy code" to keep around if needed at any stage as default finite difference-implementation. More... | |
| function fx = | evaluateComponentsMulti (pts, ends, idx, self,colvec< double > x,double t,colvec< double > mu) |
| virtual function fx = | evaluateComponents (rowvec< integer > pts,rowvec< integer > ends,rowvec< integer > idx,rowvec< integer > self,matrix< double > x,rowvec< double > t,colvec< double > mu) |
| This is the template method that actually evaluates the components at given points and values. More... | |
 Protected Member Functions inherited from KerMorObject Protected Member Functions inherited from KerMorObject | |
| function | checkType (obj, type) |
| Object typechecker. More... | |
 Protected Member Functions inherited from DPCMObject Protected Member Functions inherited from DPCMObject | |
| function | registerProps (varargin) |
| Call this method at any class that defines DPCM observed properties. More... | |
| function | registerProps (varargin) |
Additional Inherited Members | |
 Static Protected Member Functions inherited from dscomponents.ACompEvalCoreFun Static Protected Member Functions inherited from dscomponents.ACompEvalCoreFun | |
| static function obj = | loadobj (obj, from) |
 Static Protected Member Functions inherited from dscomponents.ACoreFun Static Protected Member Functions inherited from dscomponents.ACoreFun | |
| static function obj = | loadobj (obj, from) |
 Static Protected Member Functions inherited from DPCMObject Static Protected Member Functions inherited from DPCMObject | |
| static function obj = | loadobj (obj, from) |
| Re-register any registered change listeners! More... | |
| static function obj = | loadobj (obj, from) |
 Static Protected Member Functions inherited from general.AProjectable Static Protected Member Functions inherited from general.AProjectable | |
| static function obj = | loadobj (obj, from) |
 Protected Attributes inherited from dscomponents.ACompEvalCoreFun Protected Attributes inherited from dscomponents.ACompEvalCoreFun | |
| matrix< double > | S = {""} |
| The x-component selection matrices (precomputed on setting PointSet/AltPointSet). �S� is passed to the function evaluating the components of �� or its derivatives. More... | |
| models.muscle.Dynamics.Dynamics | ( | sys | ) |
Definition at line 201 of file Dynamics.m.
| function copy = models.muscle.Dynamics.clone | ( | ) |
Create new instance.
Definition at line 339 of file Dynamics.m.
| function [ SPK , SPg , SPalpha , SPLamDot ] = models.muscle.Dynamics.computeSparsityPattern | ( | ) |
Computes all sorts of patterns simultaneously.
Definition at line 20 of file computeSparsityPattern.m.
References k.
Referenced by configUpdated().

| function models.muscle.Dynamics.configUpdated | ( | ) |
Definition at line 212 of file Dynamics.m.
References computeSparsityPattern(), dscomponents.ACoreFun.fDim, dscomponents.ACoreFun.JSparsityPattern, nfibres, and dscomponents.ACoreFun.xDim.

|
protected |
Computes specified partial derivatives of \(f\) of the components given by pts and the selected partial derivatives by dfxsel.
| pts | The components of \(f\) for which derivatives are required |
| ends | At the \(i\)-th entry it contains the last position in the \(\vx\) vector that indicates an input value relevant for the \(i\)-th point evaluation, i.e. \(f_i(\vx) = f_i(\vx(ends(i-1){:}ends(i)));\) |
| idx | The indices of \(\vx\)-entries in the global \(\vx\) vector w.r.t the \(i\)-th point, e.g. \(xglobal(i-1:i+1) = \vx(ends(i-1):ends(i))\) |
| deriv | The indices within \(\vx\) that derivatives are required for. |
| self | The positions in the \(\vx\) vector that correspond to the \(i\)-th output dimension, if applicable (usually \(f_i\) depends on \(x_i\), but not necessarily) |
| x | The state space location \(\vx\) |
| t | The corresponding times \(t\) for the state \(\vx\) |
| dfxsel | A derivative selection matrix. Contains the mapping for each row of x to the output points pts. As deriv might contain less than size(x,1) values, use |
| mu | The corresponding parameter \(\mu\) for the state \(\vx\) |
| dfxsel(,deriv)' | to select the mapping for the actually computed derivatives. |
| dfx | A column vector with numel(deriv) rows containing the derivatives at all specified pts i with respect to the coordinates given by idx(ends(i-1):ends(i)) |
Definition at line 429 of file Dynamics.m.
|
protected |
This is the template method that actually evaluates the components at given points and values.
| pts | The components of \(\vf\) for which derivatives are required |
| ends | At the \(i\)-th entry it contains the last position in the \(\vx\) vector that indicates an input value relevant for the \(i\)-th point evaluation, i.e. \(f_i(\vx) = f_i(\vx(ends(i-1)+1{:}ends(i)));\) |
| unused1 | Marked as "~" in original m-file. |
| unused2 | Marked as "~" in original m-file. |
| x | A matrix \(\vX\) with the state space locations \(\vx_i\) in its columns. In rows end(i-1)+1:end(i) it contains the states relevant to evaluate the i-th component of ��. States occur multiply in �� if different components of �� depend on these states. |
| unused3 | Marked as "~" in original m-file. |
| idx | The indices of \(\vx\)-entries in the global \(\vx\) vector w.r.t the \(i\)-th point, e.g. \(xglobal(i-1:i+1) = \vx(ends(i-1):ends(i))\) |
| self | The positions in the \(\vx\) vector that correspond to the \(i\)-th output dimension, if applicable (usually \(f_i\) depends on \(x_i\), but not necessarily) |
| t | The corresponding times \(t_i\) for each state \(\vx_i\) |
| mu | The corresponding parameters \(\mu_i\) for each state \(\vx_i\), as column matrix |
| fx | A matrix with pts-many component function evaluations \(f_i(\vx)\) as rows and as many columns as \(\vX\) had. |
Definition at line 391 of file Dynamics.m.
| function fx = models.muscle.Dynamics.evaluateComponentSet | ( | integer | nr, |
| colvec< double > | x, | ||
| double | t | ||
| ) |
Computes the full or reduced component functions of the given point set.
| nr | The number of the PointSet to use. |
| x | The state space location \(\vx\) |
| t | The corresponding times \(t\) for the state \(\vx\) |
Definition at line 271 of file Dynamics.m.
References dscomponents.ACoreFun.evaluate(), dscomponents.ACompEvalCoreFun.PointSets, and general.AProjectable.V.

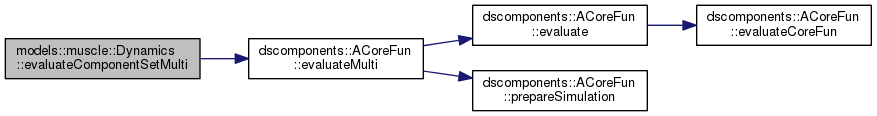
| function fx = models.muscle.Dynamics.evaluateComponentSetMulti | ( | integer | nr, |
| matrix< double > | x, | ||
| rowvec< double > | t, | ||
| matrix< double > | mu | ||
| ) |
Computes the full component functions of the given point set.
| nr | The number of the PointSet to use. |
| x | The state space locations \(\vx\) |
| t | The corresponding times \(t\) for the state \(\vx\) |
| mu | The corresponding parameters \(\mu\). Can be a single parameter or as many as the size of x |
Definition at line 288 of file Dynamics.m.
References dscomponents.ACoreFun.evaluateMulti(), and dscomponents.ACompEvalCoreFun.PointSets.
Actual method used to evaluate the dynamical sytems' core function.
Subclasses might implement this method and set the flag CustomProjection appropriately. However, for speed reasons, if both are true one might as well override the evaluate member directly as it basically cares for the cases when one of the flags is not true. In that case it is still important to set both flags to true as some components rely on them.
| x | The state variable vector \(\vx(t)\) |
| t | The current time(s) \(t \in [0, T]\) |
Implements dscomponents.ACoreFun.
Definition at line 266 of file Dynamics.m.
Definition at line 229 of file Dynamics.m.
References alpha, AnisoPassiveMuscle, AnisoPassiveMuscleDeriv, AnisoPassiveTendon, AnisoPassiveTendonDeriv, dscomponents.ACoreFun.mu, nfevals, and nJevals.
| function copy = models.muscle.Dynamics.project | ( | V, | |
| W | |||
| ) |
Definition at line 366 of file Dynamics.m.
| function models.muscle.Dynamics.setSystem | ( | sys | ) |
Definition at line 356 of file Dynamics.m.
| function res = models.muscle.Dynamics.test_Jacobian | ( | matrix< double > | y, |
| double | t, | ||
| colvec< double > | mu | ||
| ) |
Overrides the random argument jacobian test as restrictions on the possible x values (detF = 1) hold.
Currently the tests using viscosity are commented out as we assume linear damping, which is extracted as extra \(A(t,\mu)\) part in the models' system
| y | The output variable vector \(\vy(t) = \vC(t,\vmu)\vx(t),~t=0\ldots T\) (general notation) |
| t | The current time(s) \(t \in [0, T]\) |
| mu | The currently used parameter \(\vmu\). Set to \([]\) if not used. |
Definition at line 308 of file Dynamics.m.
References models.BaseSecondOrderSystem.getX0(), and models.BaseFirstOrderSystem.Model.

| models.muscle.Dynamics.alpha = @("t")0 |
The activation of the muscle at time t [-].
Default: @(t)0 @(t)0
Definition at line 46 of file Dynamics.m.
Referenced by prepareSimulation().
| models.muscle.Dynamics.AnisoPassiveMuscle |
Definition at line 100 of file Dynamics.m.
Referenced by prepareSimulation().
| models.muscle.Dynamics.AnisoPassiveMuscleDeriv |
Definition at line 102 of file Dynamics.m.
Referenced by prepareSimulation().
| models.muscle.Dynamics.AnisoPassiveTendon |
Definition at line 104 of file Dynamics.m.
Referenced by prepareSimulation().
| models.muscle.Dynamics.AnisoPassiveTendonDeriv |
Definition at line 106 of file Dynamics.m.
Referenced by prepareSimulation().
| models.muscle.Dynamics.APExp |
Definition at line 98 of file Dynamics.m.
| models.muscle.Dynamics.b1cf = 5.316372204148964 |
Cross-fibre markert part [MPa] = [N/mm²].
Default: 5.316372204148964
Definition at line 30 of file Dynamics.m.
| models.muscle.Dynamics.ComputeUnassembled = false |
% Unassembled stuff
Default: false
Definition at line 57 of file Dynamics.m.
| models.muscle.Dynamics.d1cf = 0.014991843974911 |
| models.muscle.Dynamics.fDim_unass |
Definition at line 79 of file Dynamics.m.
| models.muscle.Dynamics.ForceLengthFun |
% Force length function fields
Definition at line 86 of file Dynamics.m.
| models.muscle.Dynamics.ForceLengthFunDeriv |
Definition at line 93 of file Dynamics.m.
| models.muscle.Dynamics.idx_uv_bc_glob_unass |
The indices of any dirichlet value in the unassembled vector duvw.
Definition at line 72 of file Dynamics.m.
| models.muscle.Dynamics.idx_v_elems_unass |
Definition at line 83 of file Dynamics.m.
| models.muscle.Dynamics.lambda_dot |
Definition at line 121 of file Dynamics.m.
| models.muscle.Dynamics.lambda_dot_pos |
Helper variable for fullmodels.muscle.model.
SetAccess = Protected, GetAccess = Public Definition at line 111 of file Dynamics.m.
| models.muscle.Dynamics.LastBCResiduals |
Prepared arguments for APExpansion muprep;.
Transient set to true. SetAccess = Private, GetAccess = Public Definition at line 129 of file Dynamics.m.
| models.muscle.Dynamics.nfevals |
Definition at line 142 of file Dynamics.m.
Referenced by prepareSimulation().
| models.muscle.Dynamics.nfibres |
Definition at line 124 of file Dynamics.m.
Referenced by configUpdated().
| models.muscle.Dynamics.nJevals |
Definition at line 144 of file Dynamics.m.
Referenced by prepareSimulation().
| models.muscle.Dynamics.num_dv_unass |
Definition at line 81 of file Dynamics.m.
| models.muscle.Dynamics.RampTime |
Helper value for QuickReleaseTests (or others) that use a function handle with certain alpha ramp time for different simulations. Used in getSimCacheExtra to uniquely identify a simulation in the cache.
Transient set to true. Definition at line 149 of file Dynamics.m.
| models.muscle.Dynamics.Sigma |
Sigma assembly matrix.
Definition at line 65 of file Dynamics.m.